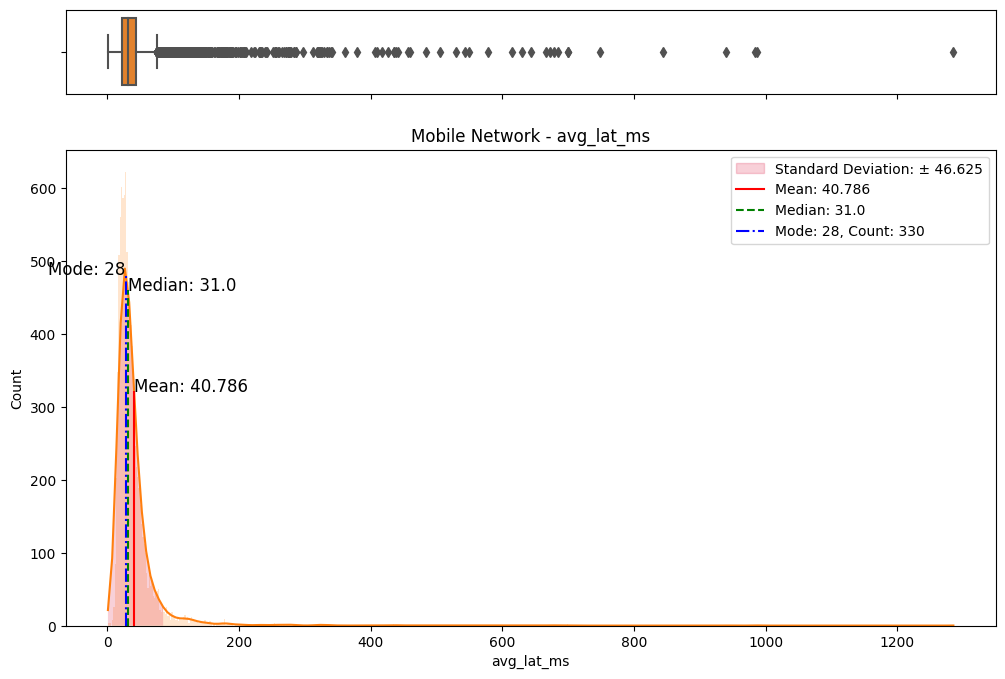
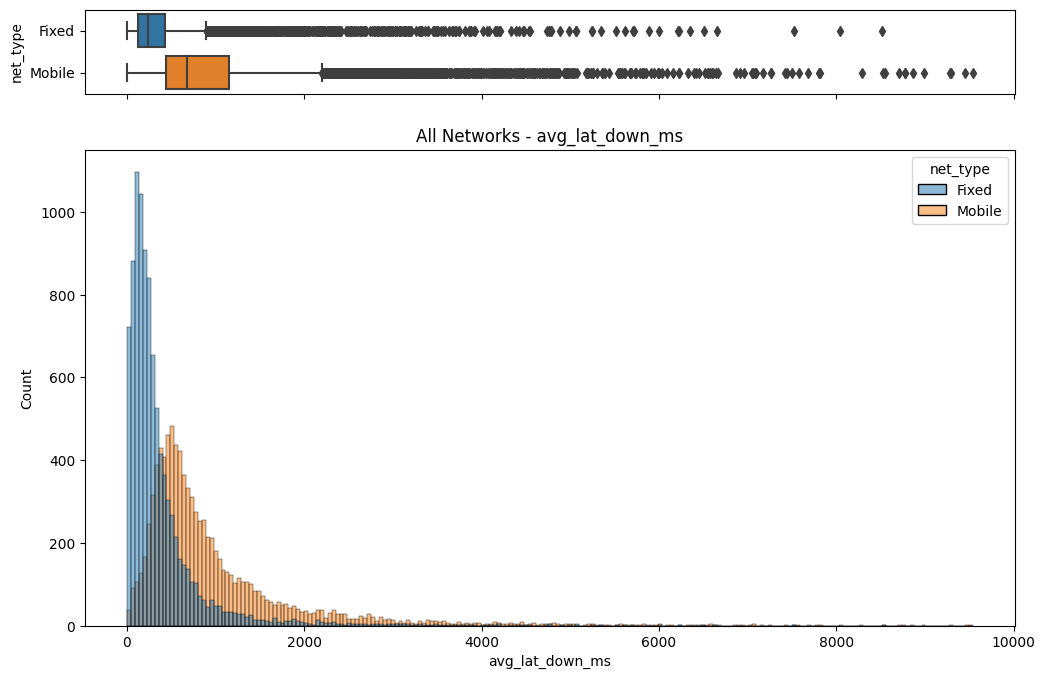
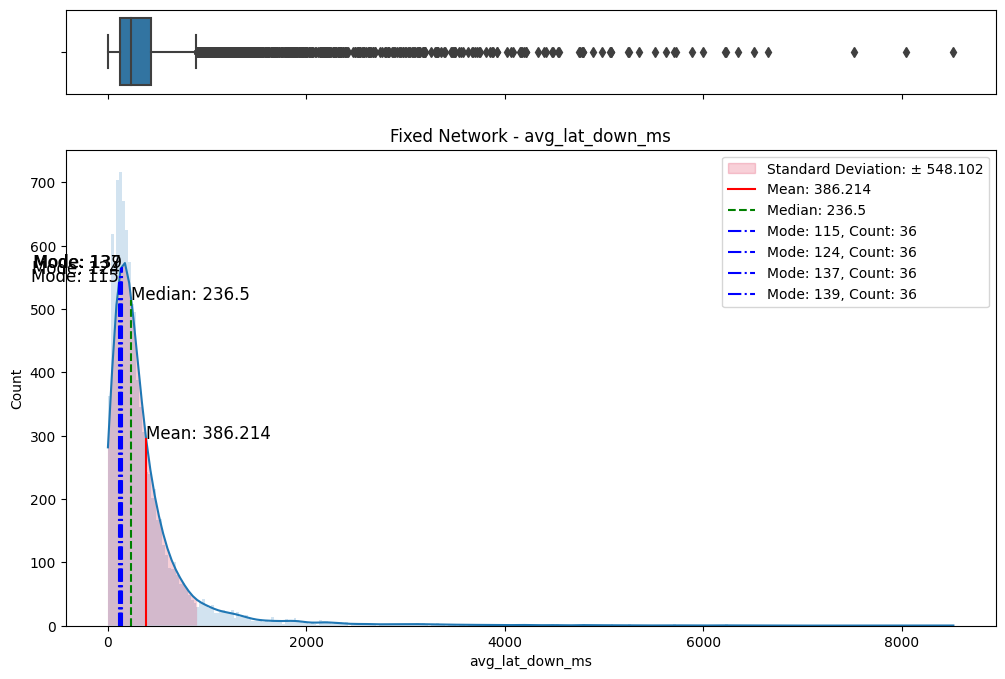
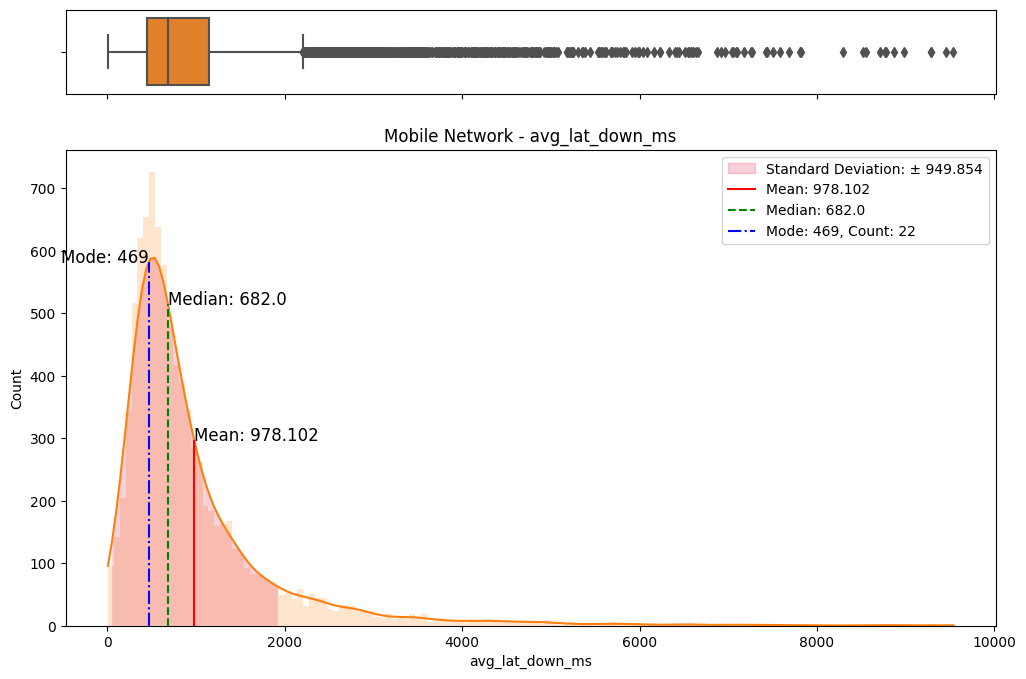
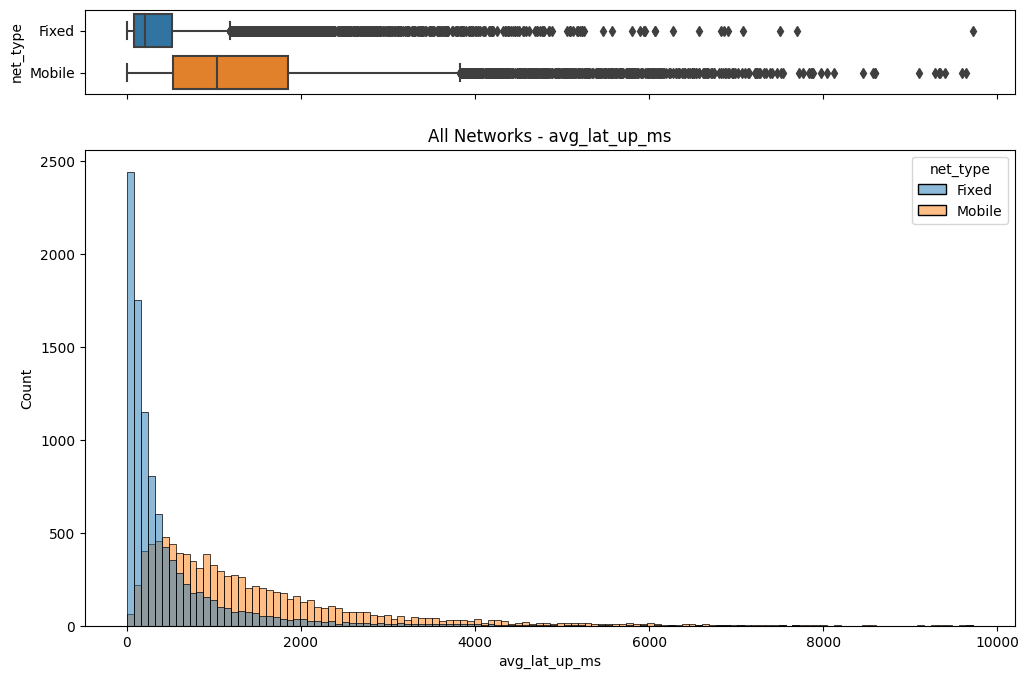
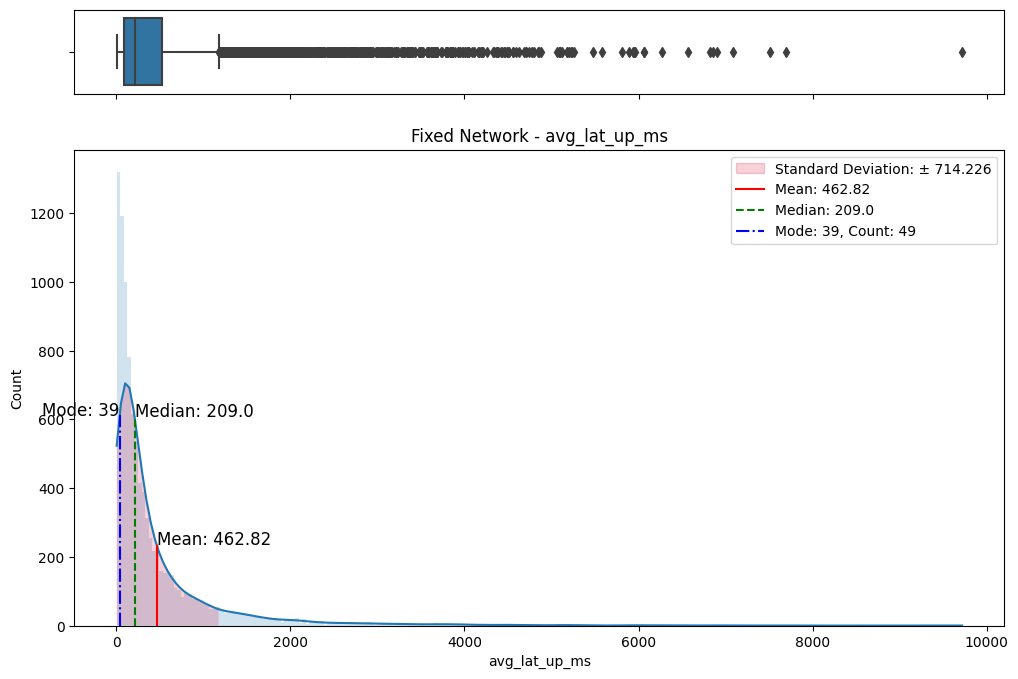
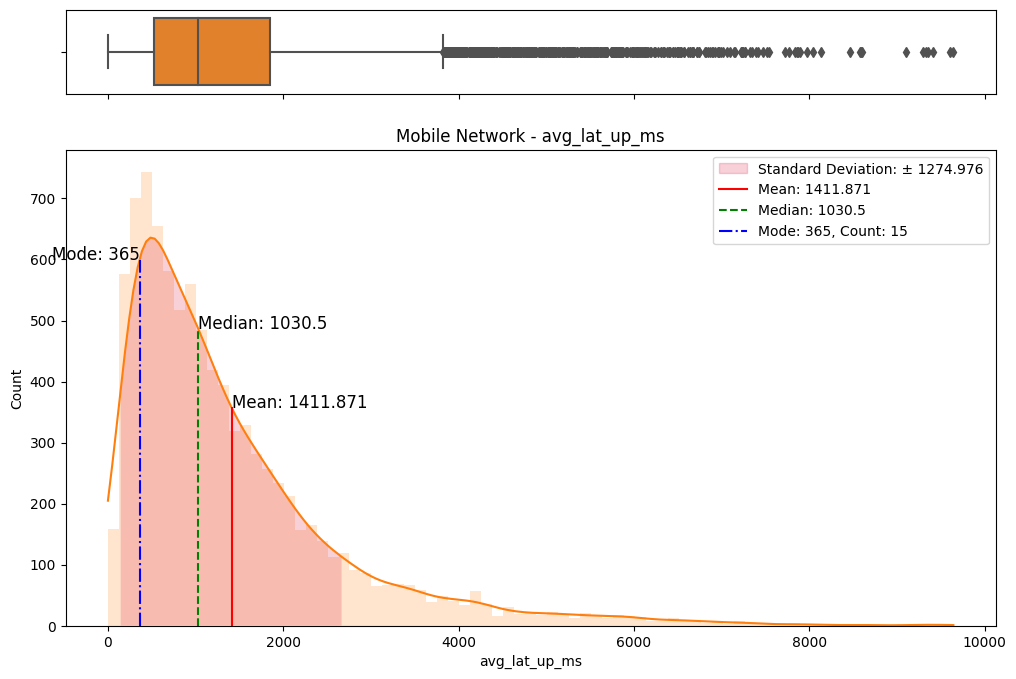
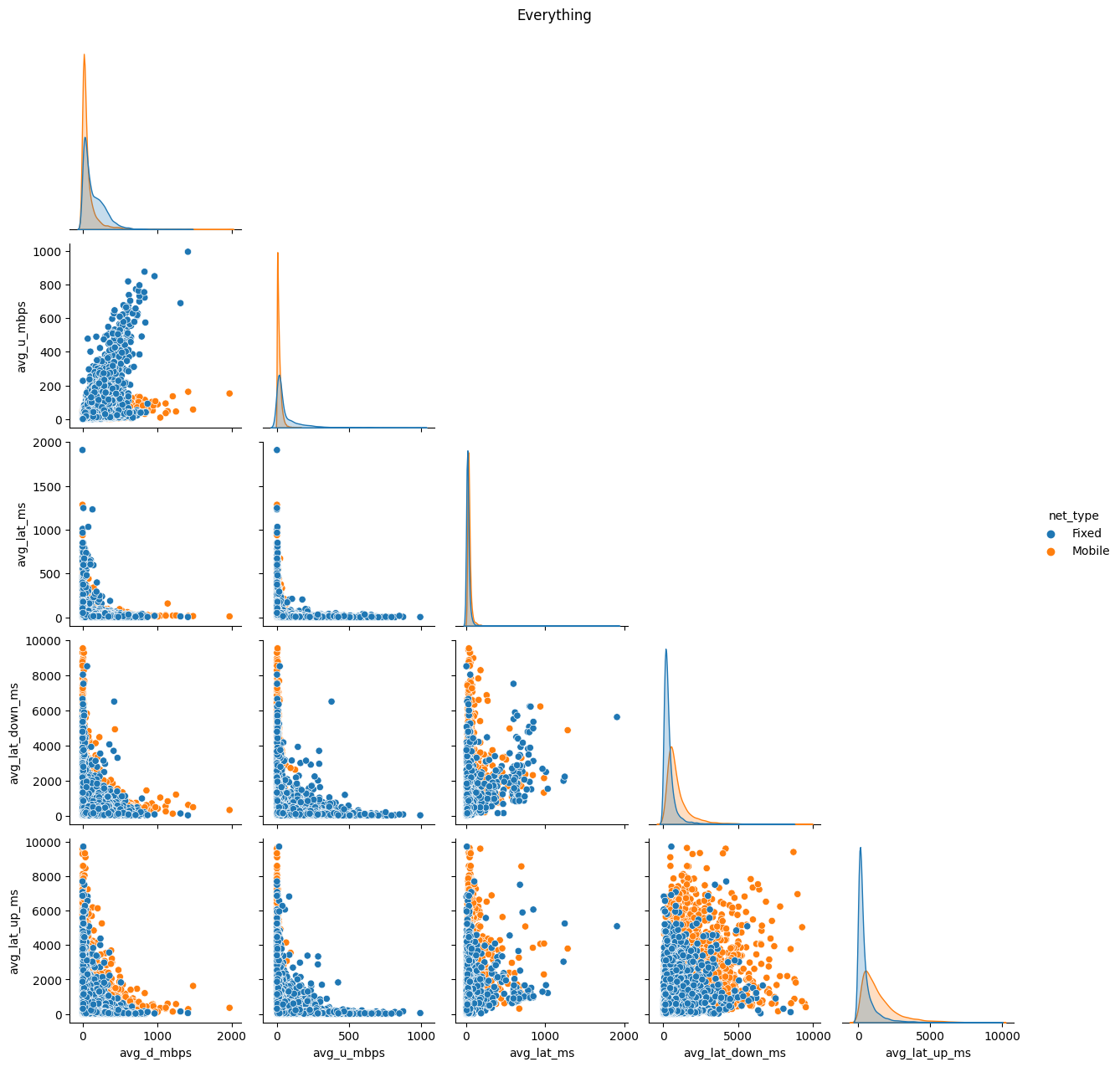
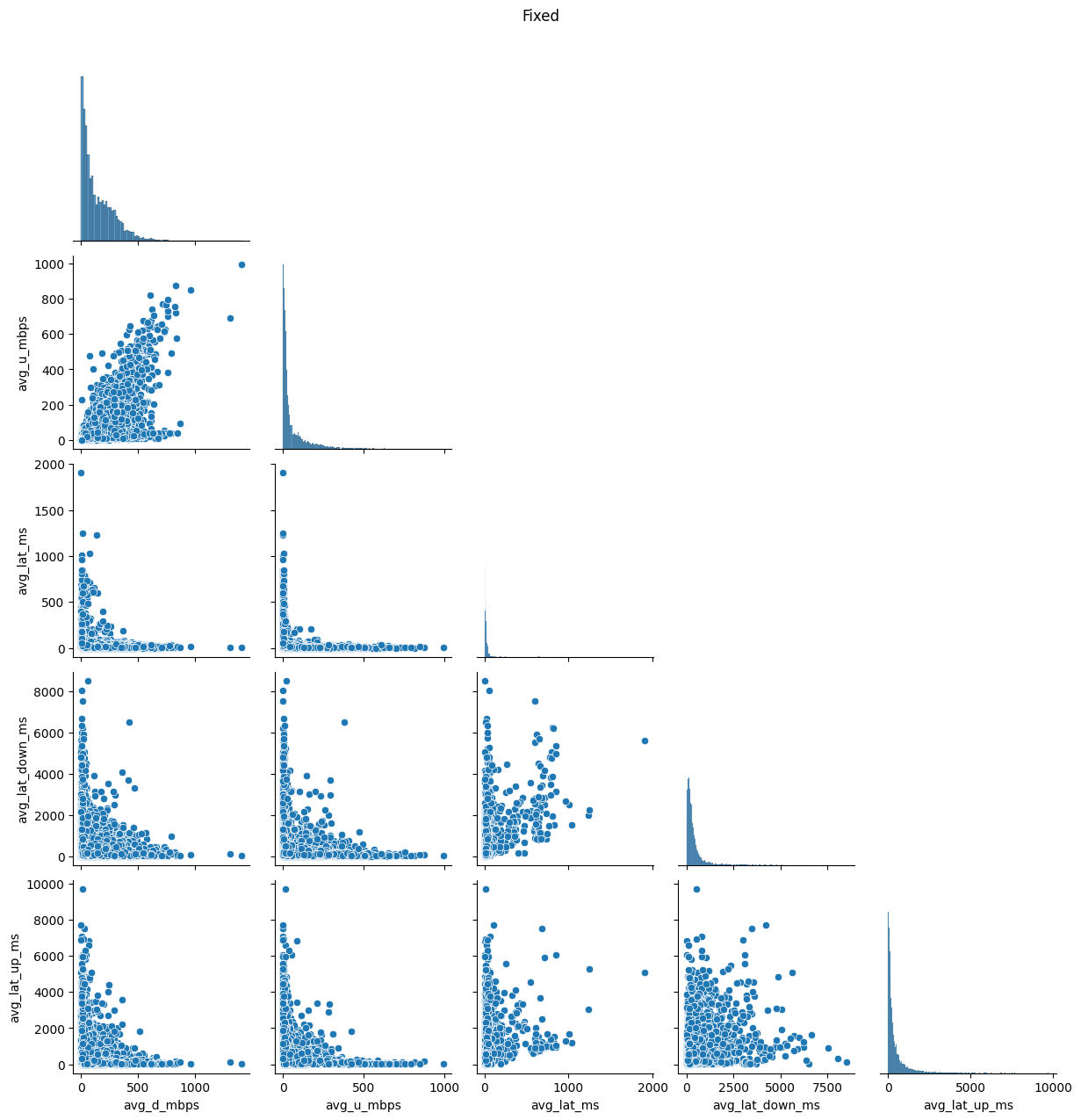
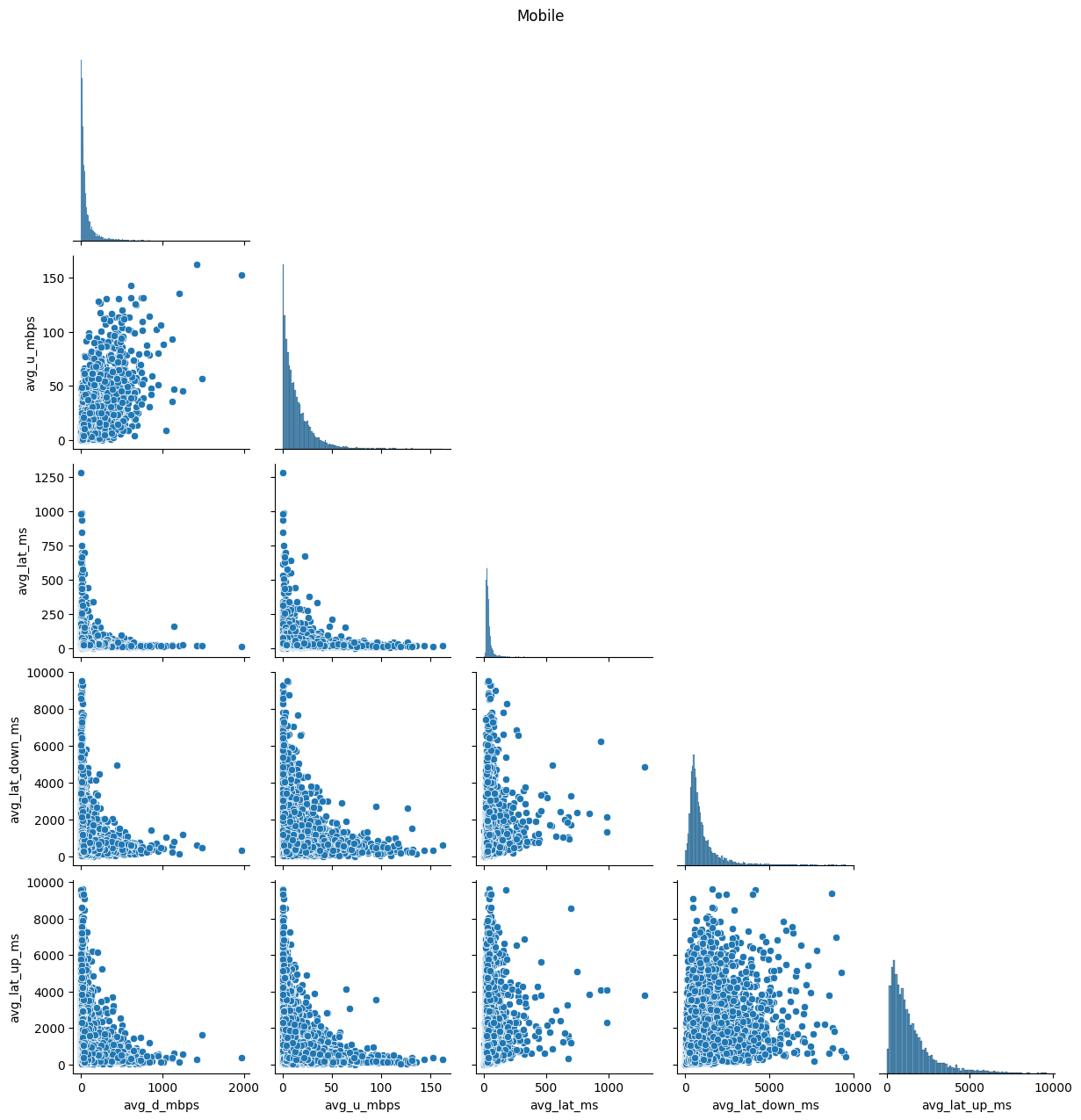
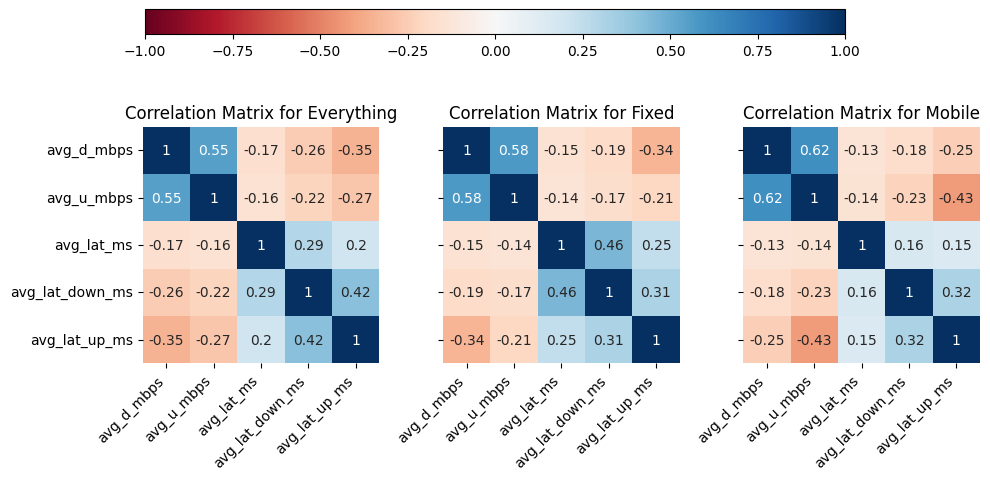
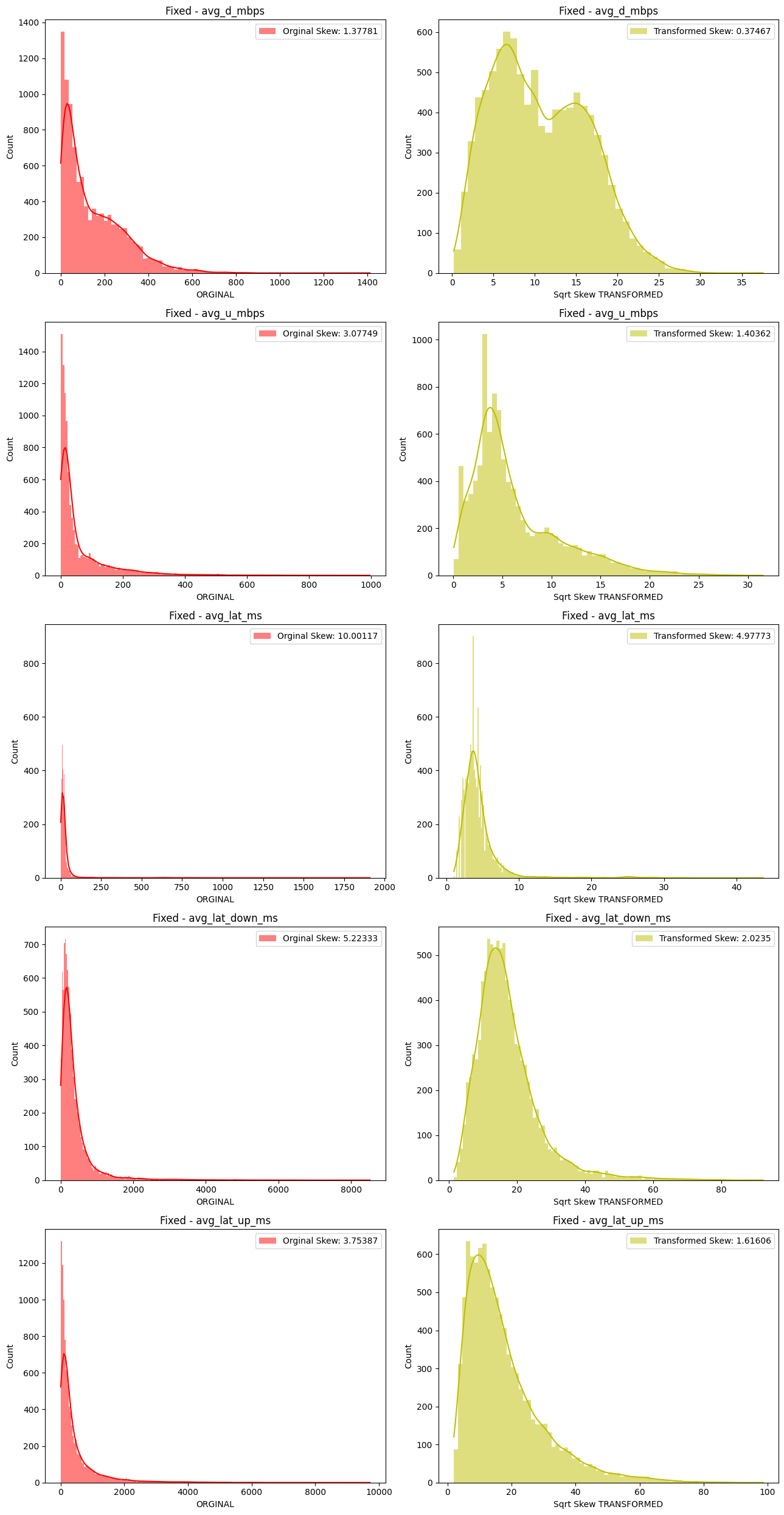
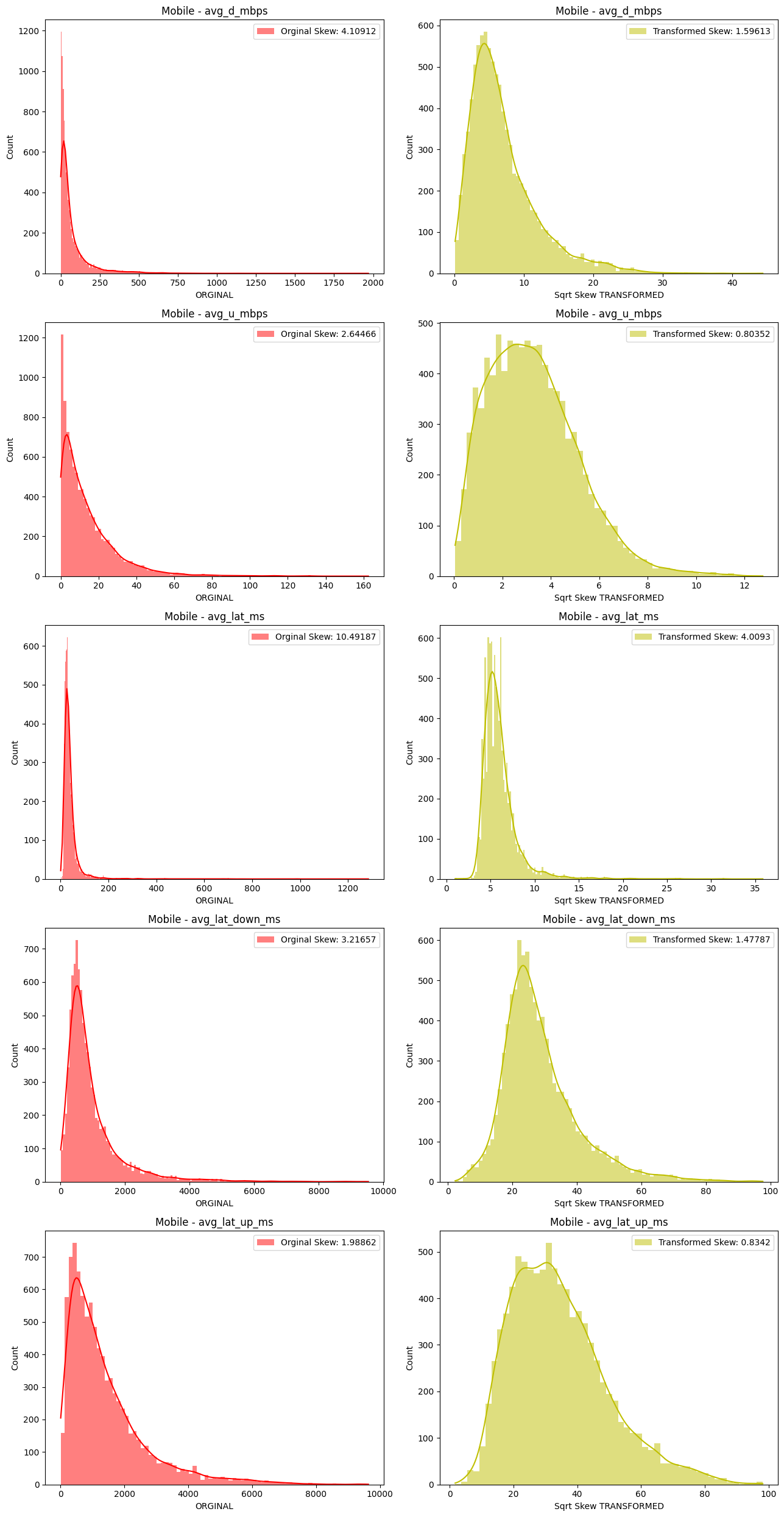
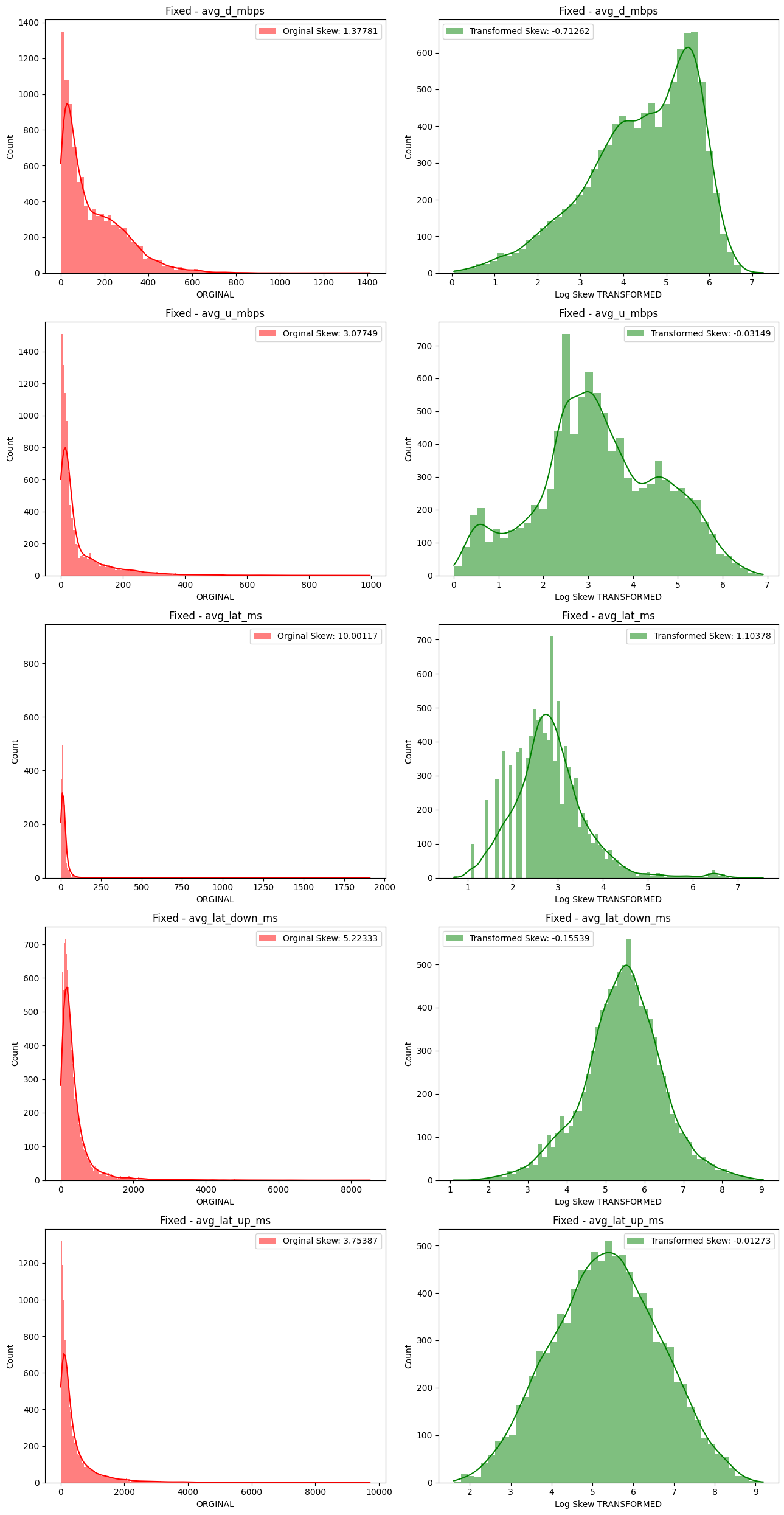
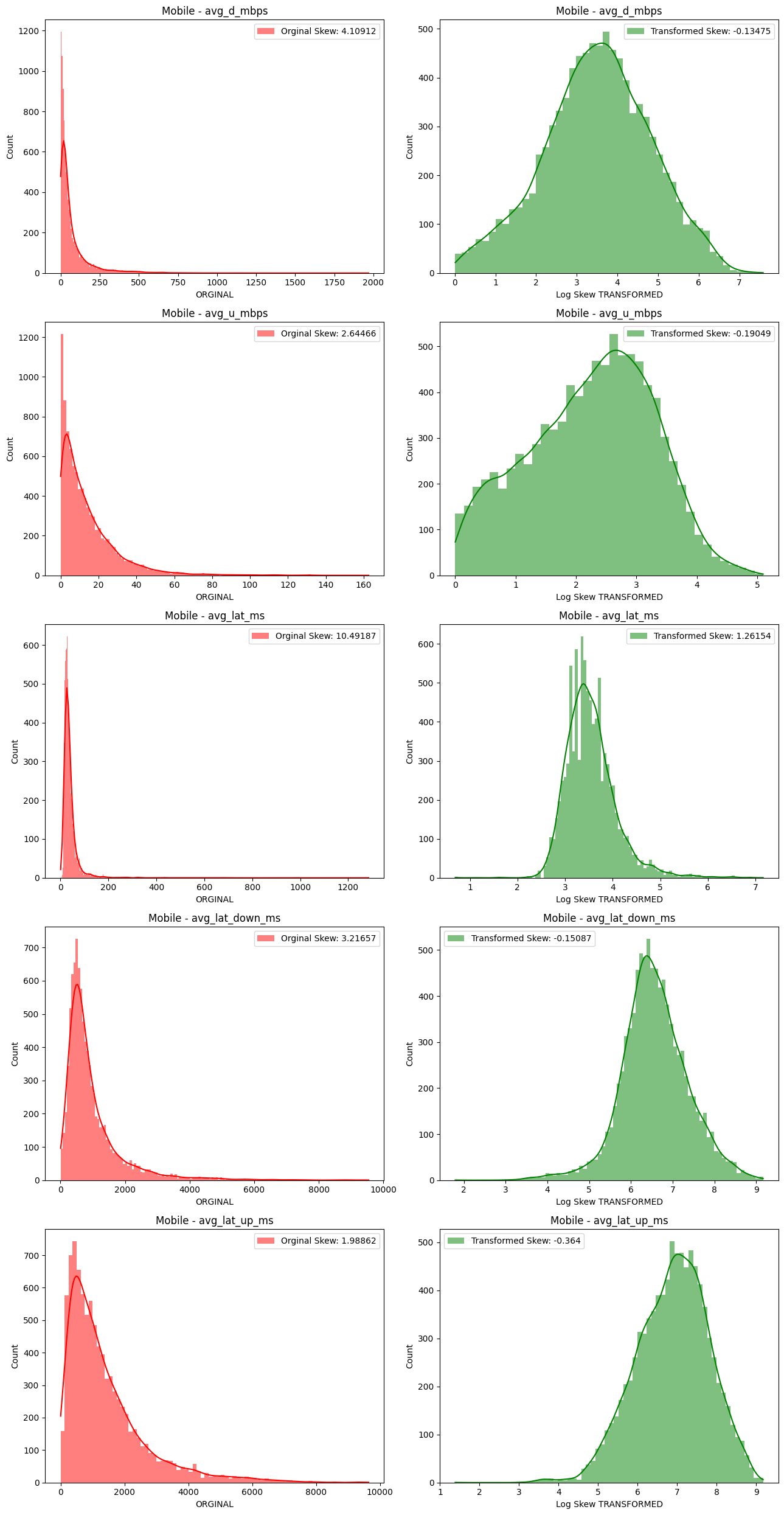
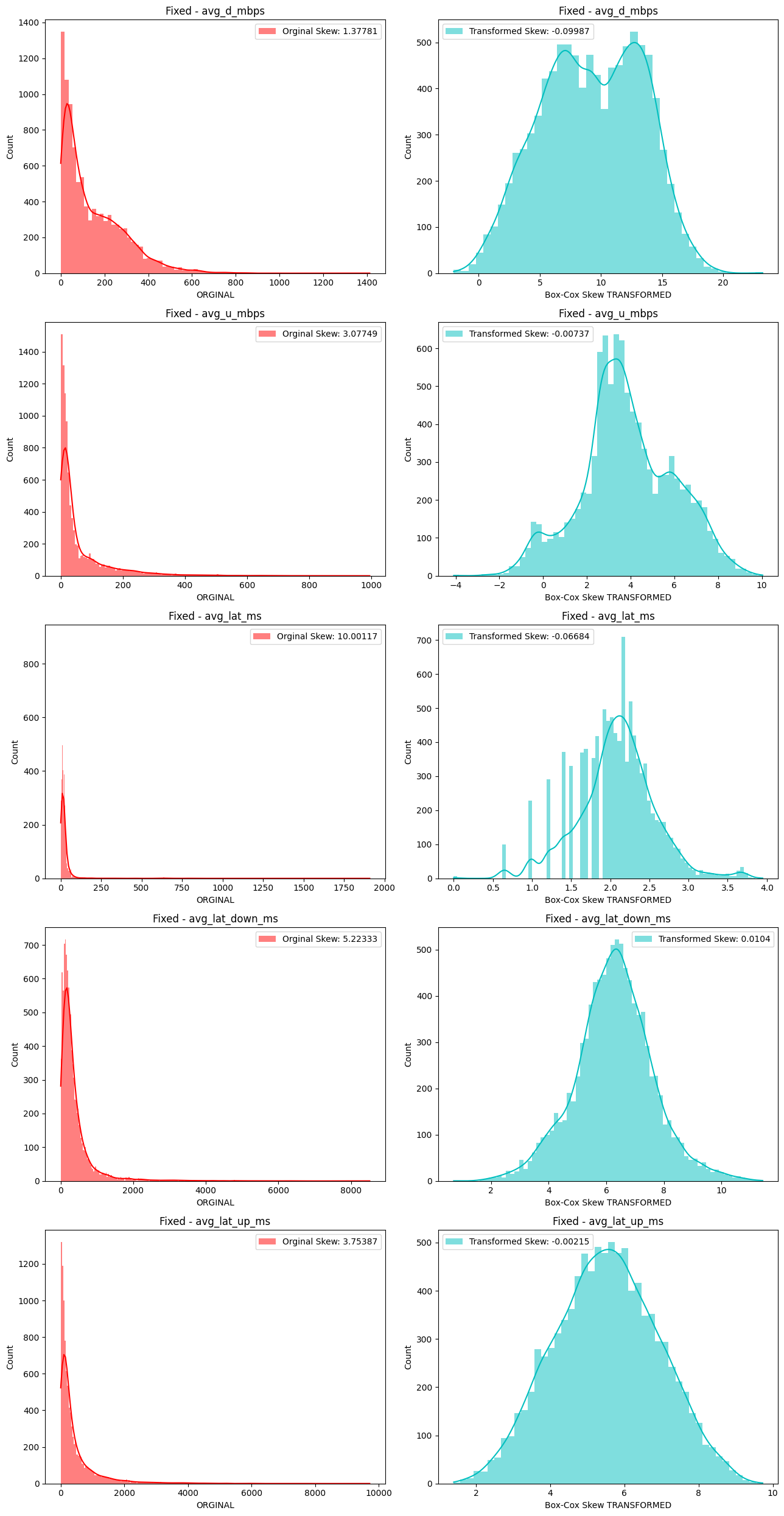
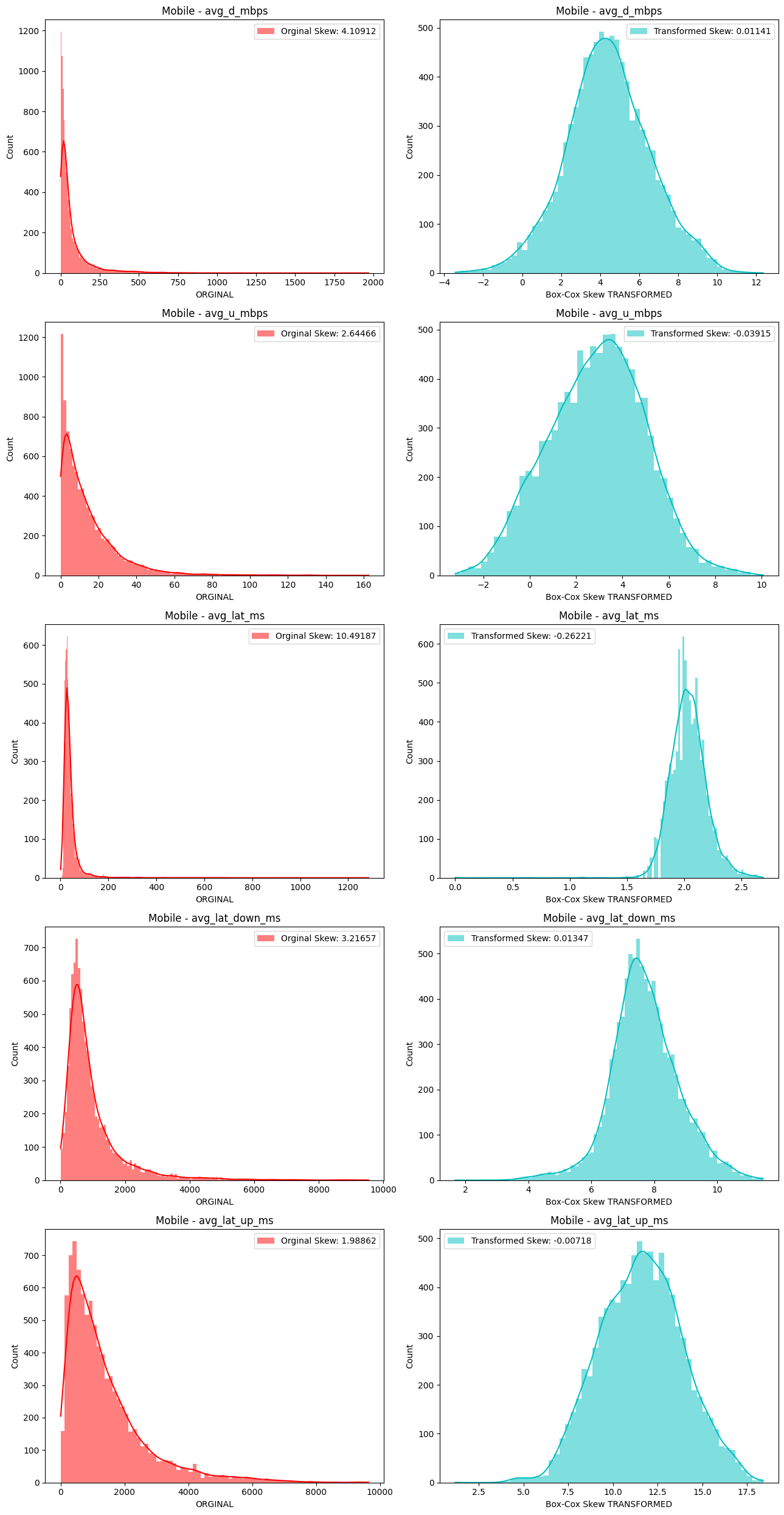
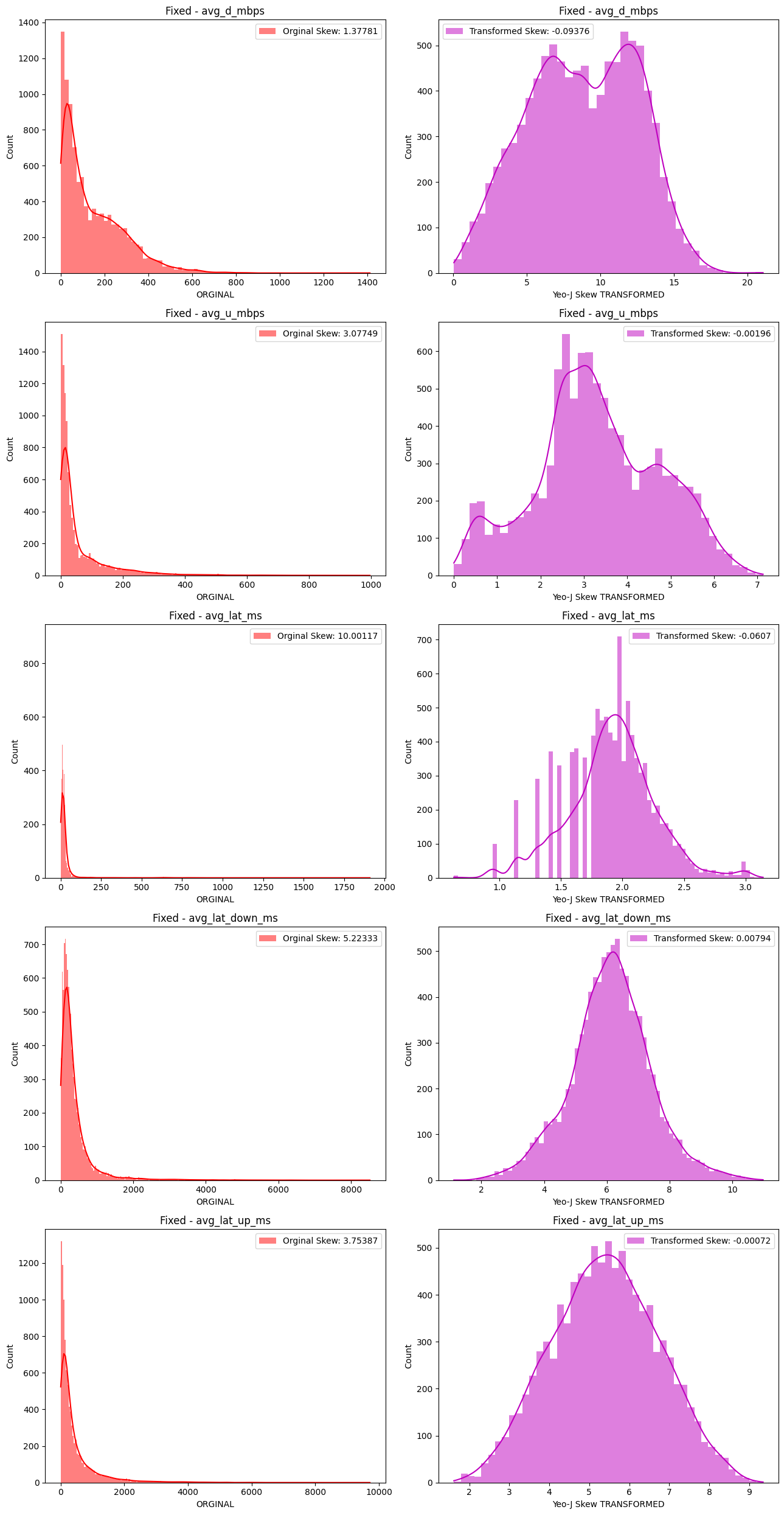
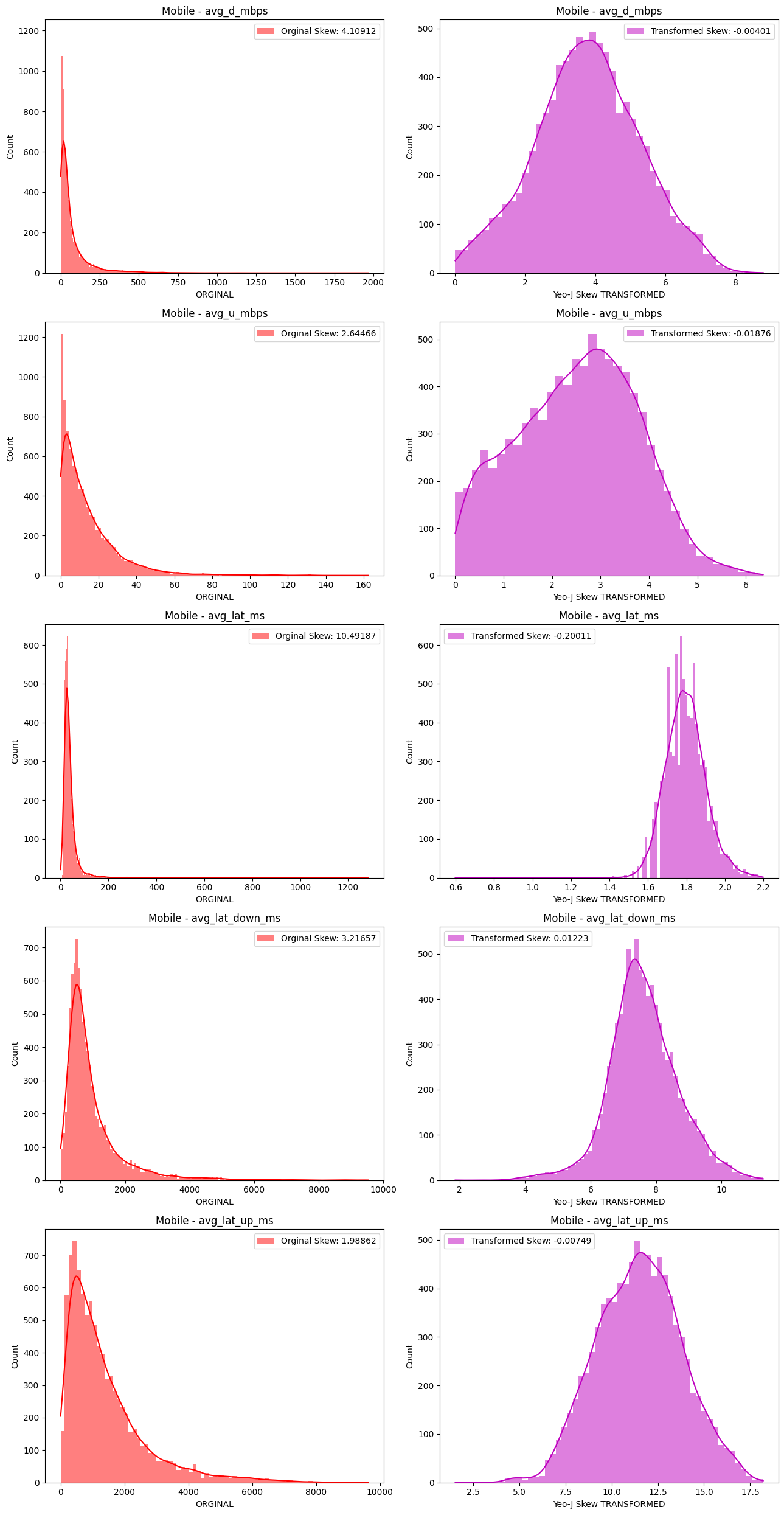
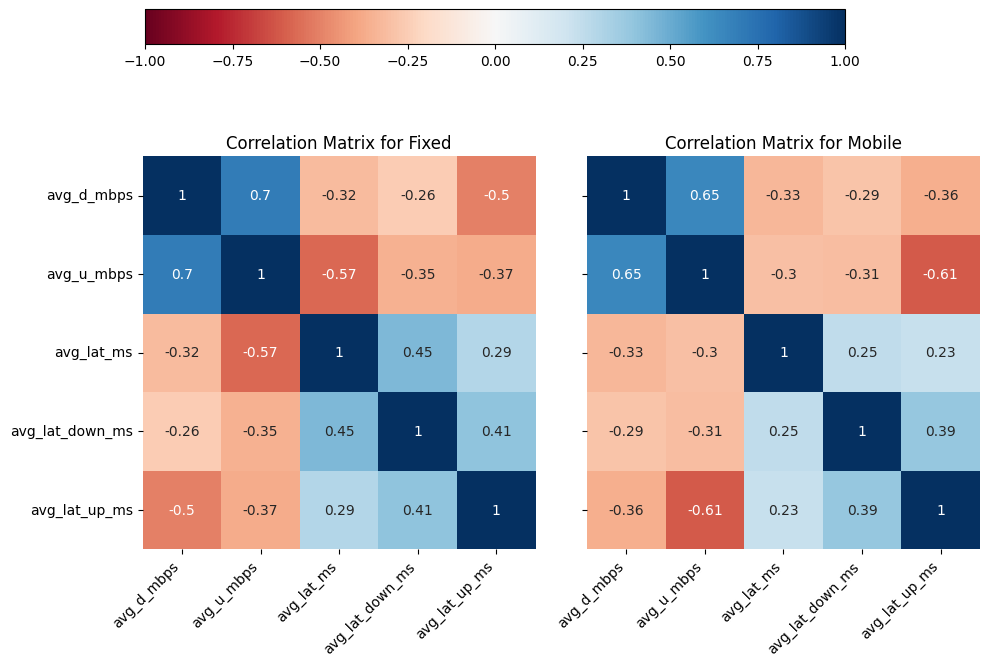
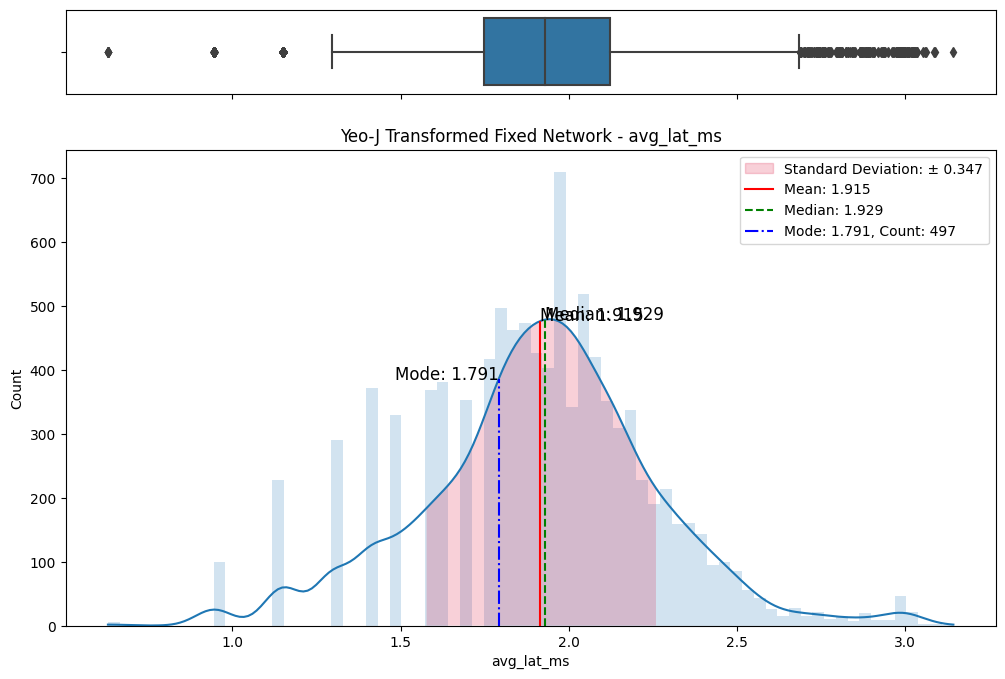
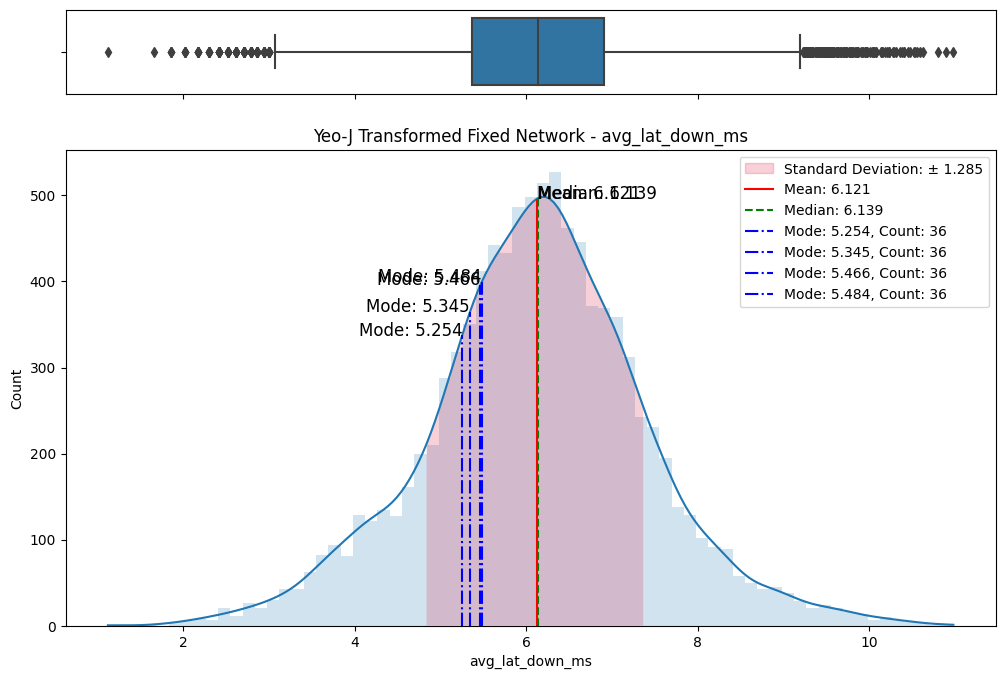
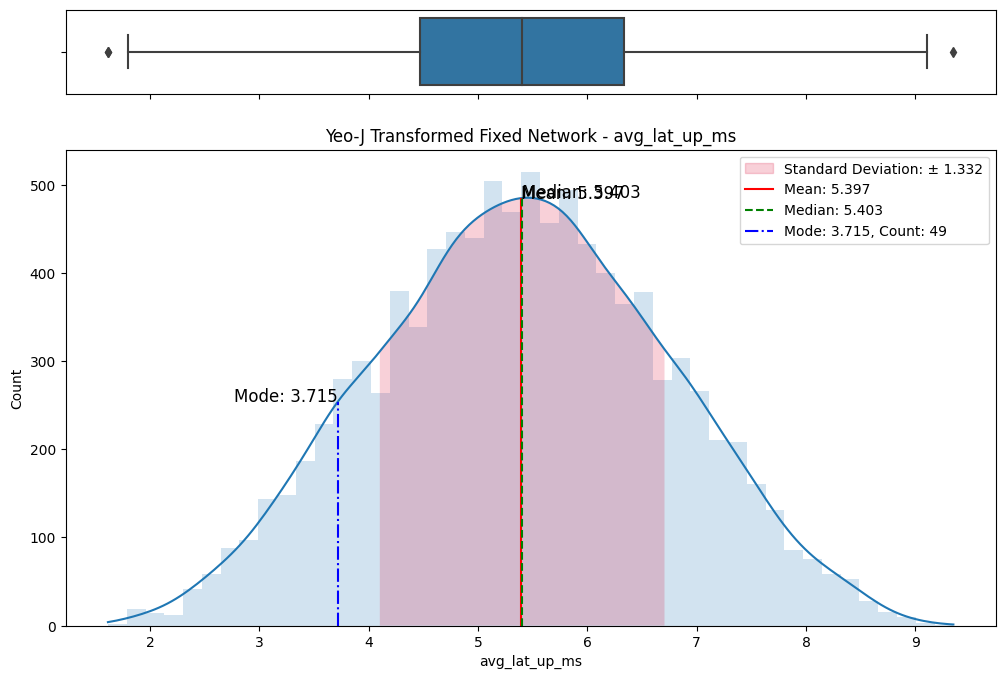
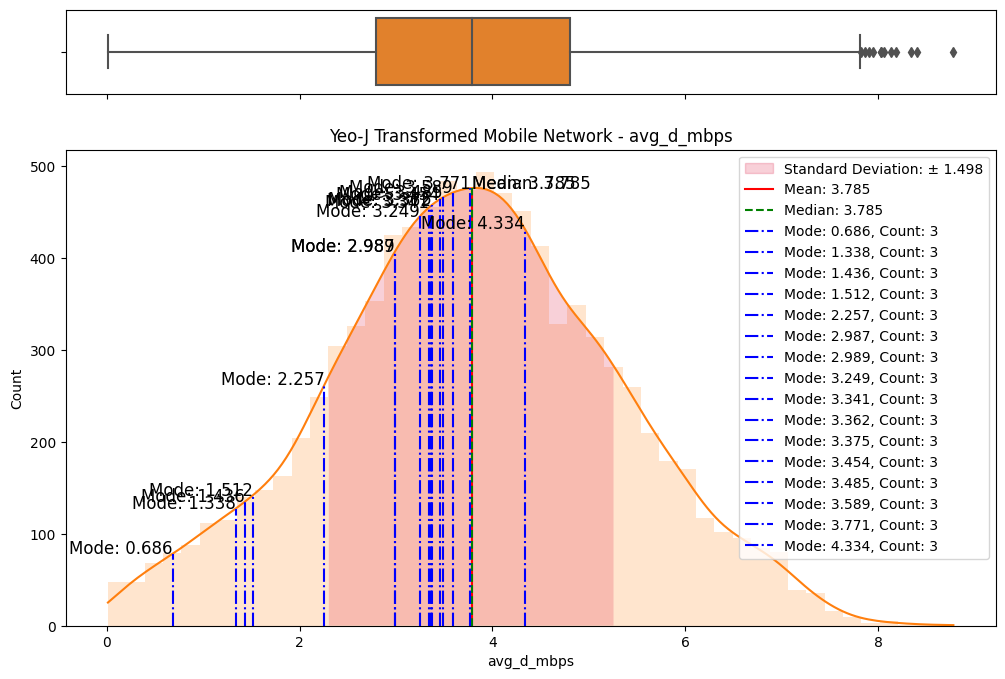
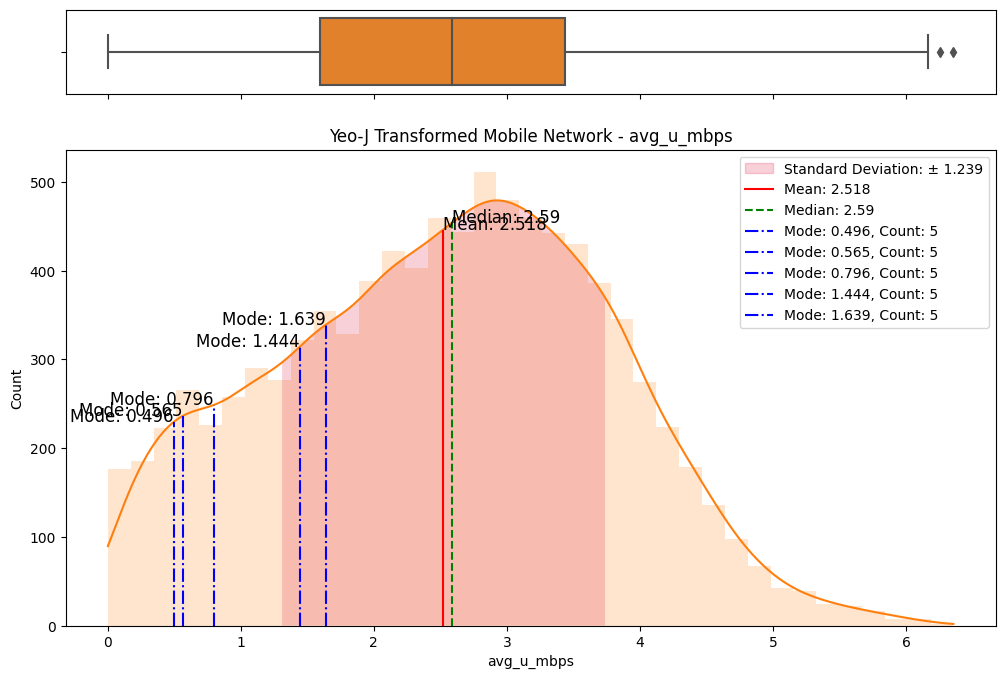
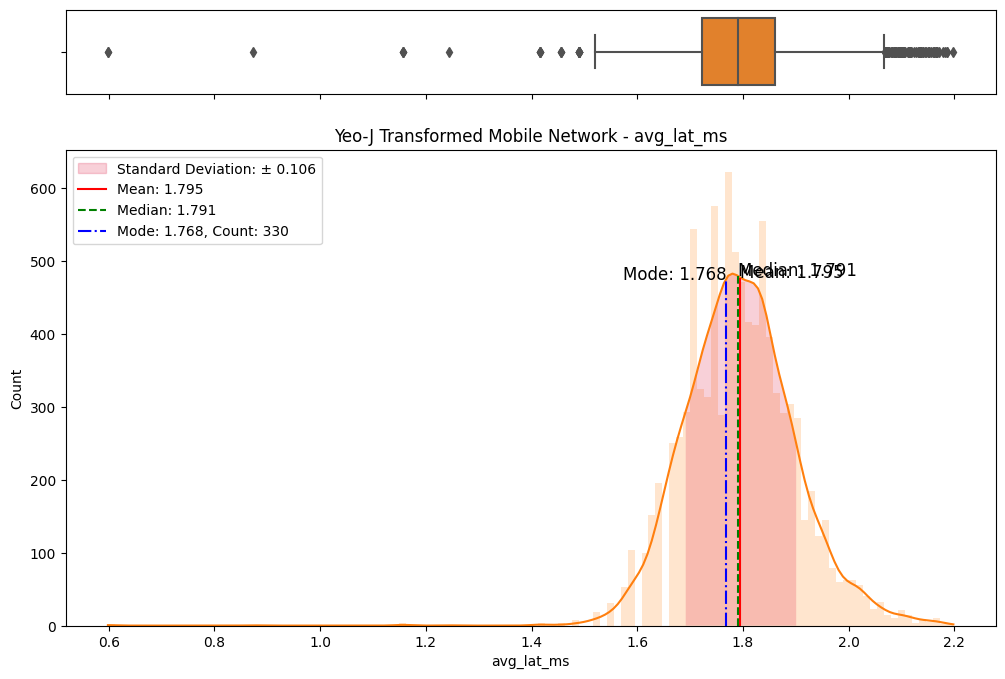
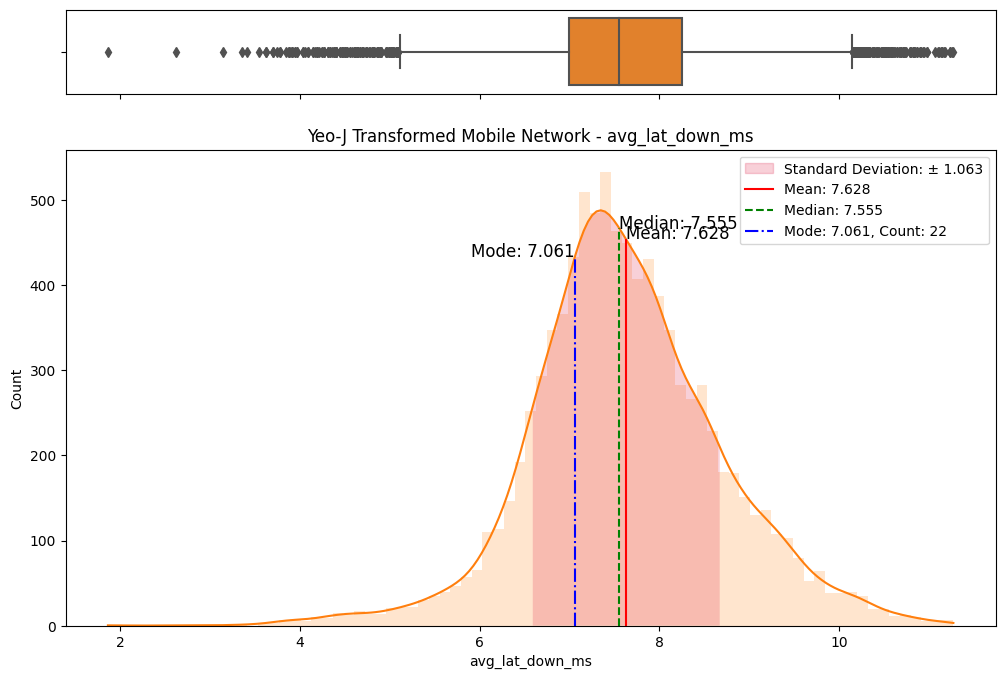
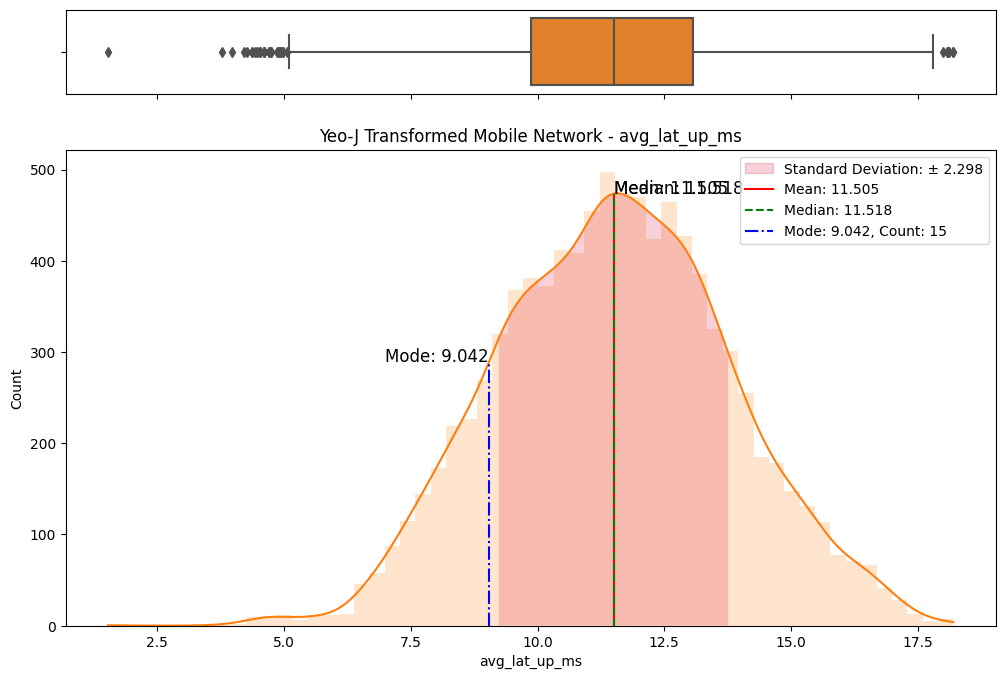
# Import dependencies
import pandas as pd
import numpy as np
from scipy import stats
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import matplotlib.lines as mlines
from matplotlib.colors import Normalize
import seaborn as sns
import plotly.figure_factory as ff
# Filter out seaborn warnings on the histplots
import warnings
warnings.filterwarnings('ignore', category=FutureWarning)
#import cleaned dataset
df = pd.read_parquet('cleaned_dataset.parquet')
# split df into two
#dataframe with entries just for the fixed category
df_fixed = df.loc[df['net_type'] == 'Fixed'].copy()
df_fixed.reset_index(drop=True, inplace=True)
df_fixed.drop(columns='net_type', inplace=True)
#dataframe with entries just for the mobile category
df_mob = df.loc[df['net_type'] == 'Mobile'].copy()
df_mob.reset_index(drop=True, inplace=True)
df_mob.drop(columns='net_type', inplace=True)
display(df)
print(df.columns)
# create dataframe lis to iterate through
dfs = {
'Fixed': [ df_fixed, 'tab:blue' ],
'Mobile': [ df_mob, 'tab:orange' ]
}
for key in dfs:
display(dfs[key][0])
print(dfs[key][0].columns)
# Check for infinite values
inf_values = np.isinf(dfs[key][0]).sum().sum()
print(f'There are {inf_values} infinite values in the {key} DataFrame.')
# Check for infinite values
inf_values = np.isinf(df.drop(columns='net_type')).sum().sum()
print(f'There are {inf_values} infinite values in the DataFrame.')| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | net_type | |
|---|---|---|---|---|---|---|
| 0 | 50.073 | 18.199 | 40 | 475 | 1954 | Mobile |
| 1 | 21.784 | 0.745 | 47 | 1493 | 2252 | Mobile |
| 2 | 18.159 | 1.662 | 21 | 244 | 2067 | Mobile |
| 3 | 1.439 | 0.659 | 749 | 2357 | 5083 | Mobile |
| 4 | 13.498 | 3.525 | 37 | 598 | 1023 | Mobile |
| ... | ... | ... | ... | ... | ... | ... |
| 19025 | 215.644 | 114.035 | 14 | 384 | 606 | Fixed |
| 19026 | 48.533 | 17.553 | 34 | 172 | 43 | Fixed |
| 19027 | 5.732 | 0.473 | 52 | 8039 | 304 | Fixed |
| 19028 | 116.025 | 129.465 | 8 | 91 | 219 | Fixed |
| 19029 | 145.911 | 42.130 | 15 | 139 | 555 | Fixed |
19030 rows × 6 columns
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms', 'net_type'],
dtype='object')
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms'],
dtype='object')
There are 0 infinite values in the Fixed DataFrame.
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms'],
dtype='object')
There are 0 infinite values in the Mobile DataFrame.
There are 0 infinite values in the DataFrame.| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | |
|---|---|---|---|---|---|
| 0 | 104.961 | 104.419 | 6 | 126 | 94 |
| 1 | 212.782 | 33.322 | 26 | 122 | 223 |
| 2 | 109.832 | 9.109 | 18 | 211 | 164 |
| 3 | 194.682 | 116.727 | 20 | 279 | 93 |
| 4 | 151.912 | 13.325 | 19 | 174 | 454 |
| ... | ... | ... | ... | ... | ... |
| 9809 | 215.644 | 114.035 | 14 | 384 | 606 |
| 9810 | 48.533 | 17.553 | 34 | 172 | 43 |
| 9811 | 5.732 | 0.473 | 52 | 8039 | 304 |
| 9812 | 116.025 | 129.465 | 8 | 91 | 219 |
| 9813 | 145.911 | 42.130 | 15 | 139 | 555 |
9814 rows × 5 columns
| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | |
|---|---|---|---|---|---|
| 0 | 50.073 | 18.199 | 40 | 475 | 1954 |
| 1 | 21.784 | 0.745 | 47 | 1493 | 2252 |
| 2 | 18.159 | 1.662 | 21 | 244 | 2067 |
| 3 | 1.439 | 0.659 | 749 | 2357 | 5083 |
| 4 | 13.498 | 3.525 | 37 | 598 | 1023 |
| ... | ... | ... | ... | ... | ... |
| 9211 | 42.572 | 23.439 | 22 | 238 | 640 |
| 9212 | 15.952 | 0.256 | 39 | 1189 | 1083 |
| 9213 | 107.443 | 25.328 | 24 | 751 | 1555 |
| 9214 | 26.593 | 21.297 | 36 | 565 | 378 |
| 9215 | 23.803 | 4.061 | 26 | 284 | 1020 |
9216 rows × 5 columns