# Import dependencies
import pandas as pd
import numpy as np
from scipy import stats
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import GradientBoostingRegressor, RandomForestRegressor, RandomForestClassifier
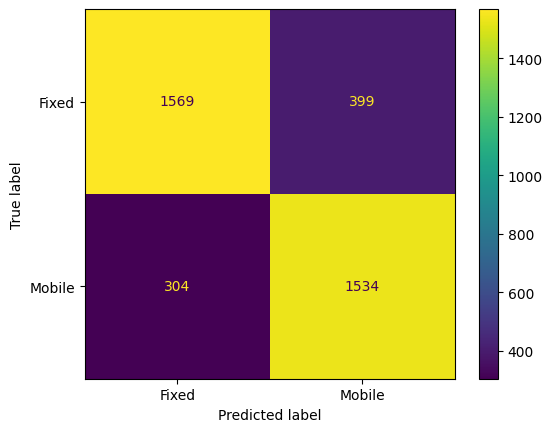
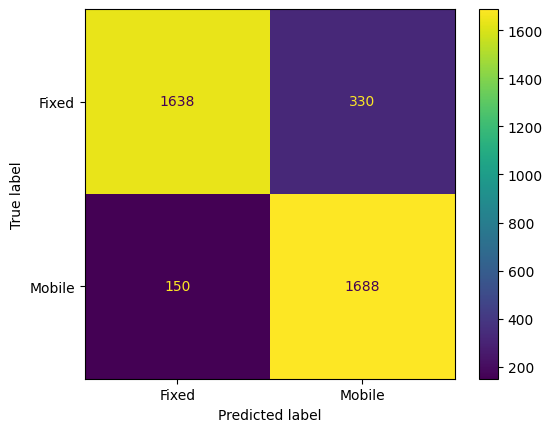
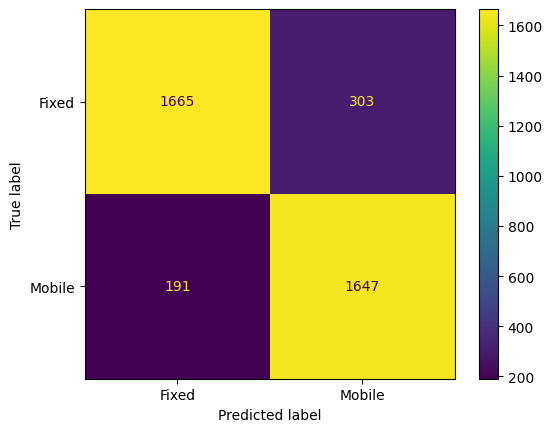
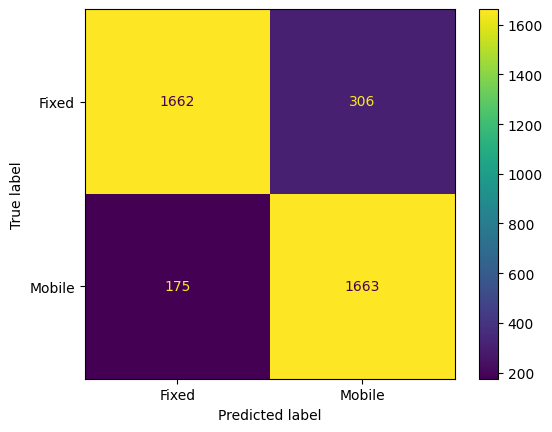
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score, classification_report, confusion_matrix, ConfusionMatrixDisplay
from sklearn.preprocessing import PowerTransformer, StandardScaler, RobustScaler
from sklearn.pipeline import Pipeline
from sklearn import svm
import plotly.figure_factory as ff
import matplotlib.pyplot as plt
#import cleaned dataset
df = pd.read_parquet('cleaned_dataset.parquet')
# split df into two
#dataframe with entries just for the fixed category
df_fixed = df.loc[df['net_type'] == 'Fixed'].copy()
df_fixed.reset_index(drop=True, inplace=True)
df_fixed.drop(columns='net_type', inplace=True)
#dataframe with entries just for the mobile category
df_mob = df.loc[df['net_type'] == 'Mobile'].copy()
df_mob.reset_index(drop=True, inplace=True)
df_mob.drop(columns='net_type', inplace=True)
display(df)
print(df.columns)
# create dataframe dict to iterate through
dfs = {
'Fixed': df_fixed,
'Mobile': df_mob,
# import YeoJ transformed dfs
'YeoJ_Fix': pd.read_parquet('yeoj_fixed_dataset.parquet'),
'YeoJ_Mob': pd.read_parquet('yeoj_mob_dataset.parquet')
}
for key in dfs:
display(dfs[key])
print(dfs[key].columns)| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | net_type | |
|---|---|---|---|---|---|---|
| 0 | 50.073 | 18.199 | 40 | 475 | 1954 | Mobile |
| 1 | 21.784 | 0.745 | 47 | 1493 | 2252 | Mobile |
| 2 | 18.159 | 1.662 | 21 | 244 | 2067 | Mobile |
| 3 | 1.439 | 0.659 | 749 | 2357 | 5083 | Mobile |
| 4 | 13.498 | 3.525 | 37 | 598 | 1023 | Mobile |
| ... | ... | ... | ... | ... | ... | ... |
| 19025 | 215.644 | 114.035 | 14 | 384 | 606 | Fixed |
| 19026 | 48.533 | 17.553 | 34 | 172 | 43 | Fixed |
| 19027 | 5.732 | 0.473 | 52 | 8039 | 304 | Fixed |
| 19028 | 116.025 | 129.465 | 8 | 91 | 219 | Fixed |
| 19029 | 145.911 | 42.130 | 15 | 139 | 555 | Fixed |
19030 rows × 6 columns
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms', 'net_type'],
dtype='object')
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms'],
dtype='object')
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms'],
dtype='object')
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms'],
dtype='object')
Index(['avg_d_mbps', 'avg_u_mbps', 'avg_lat_ms', 'avg_lat_down_ms',
'avg_lat_up_ms'],
dtype='object')| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | |
|---|---|---|---|---|---|
| 0 | 104.961 | 104.419 | 6 | 126 | 94 |
| 1 | 212.782 | 33.322 | 26 | 122 | 223 |
| 2 | 109.832 | 9.109 | 18 | 211 | 164 |
| 3 | 194.682 | 116.727 | 20 | 279 | 93 |
| 4 | 151.912 | 13.325 | 19 | 174 | 454 |
| ... | ... | ... | ... | ... | ... |
| 9809 | 215.644 | 114.035 | 14 | 384 | 606 |
| 9810 | 48.533 | 17.553 | 34 | 172 | 43 |
| 9811 | 5.732 | 0.473 | 52 | 8039 | 304 |
| 9812 | 116.025 | 129.465 | 8 | 91 | 219 |
| 9813 | 145.911 | 42.130 | 15 | 139 | 555 |
9814 rows × 5 columns
| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | |
|---|---|---|---|---|---|
| 0 | 50.073 | 18.199 | 40 | 475 | 1954 |
| 1 | 21.784 | 0.745 | 47 | 1493 | 2252 |
| 2 | 18.159 | 1.662 | 21 | 244 | 2067 |
| 3 | 1.439 | 0.659 | 749 | 2357 | 5083 |
| 4 | 13.498 | 3.525 | 37 | 598 | 1023 |
| ... | ... | ... | ... | ... | ... |
| 9211 | 42.572 | 23.439 | 22 | 238 | 640 |
| 9212 | 15.952 | 0.256 | 39 | 1189 | 1083 |
| 9213 | 107.443 | 25.328 | 24 | 751 | 1555 |
| 9214 | 26.593 | 21.297 | 36 | 565 | 378 |
| 9215 | 23.803 | 4.061 | 26 | 284 | 1020 |
9216 rows × 5 columns
| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | |
|---|---|---|---|---|---|
| 0 | 8.972164 | 4.759714 | 1.500592 | 5.364215 | 4.594474 |
| 1 | 11.504014 | 3.594225 | 2.152917 | 5.325130 | 5.469041 |
| 2 | 9.121034 | 2.338349 | 2.006061 | 5.997121 | 5.157019 |
| 3 | 11.159330 | 4.875059 | 2.049368 | 6.346418 | 4.583702 |
| 4 | 10.238558 | 2.695041 | 2.028408 | 5.758639 | 6.193776 |
| ... | ... | ... | ... | ... | ... |
| 9809 | 11.556506 | 4.850887 | 1.898827 | 6.751223 | 6.489121 |
| 9810 | 6.696179 | 2.960429 | 2.252471 | 5.744407 | 3.812195 |
| 9811 | 2.457252 | 0.387996 | 2.397371 | 10.894972 | 5.784466 |
| 9812 | 9.303431 | 4.982478 | 1.641398 | 4.972823 | 5.450639 |
| 9813 | 10.094487 | 3.830499 | 1.928811 | 5.483558 | 6.399159 |
9814 rows × 5 columns
| avg_d_mbps | avg_u_mbps | avg_lat_ms | avg_lat_down_ms | avg_lat_up_ms | |
|---|---|---|---|---|---|
| 0 | 4.239305 | 3.351656 | 1.842729 | 7.078103 | 13.227949 |
| 1 | 3.317389 | 0.569910 | 1.873058 | 8.614639 | 13.627984 |
| 2 | 3.123101 | 1.020246 | 1.700411 | 6.220586 | 13.385472 |
| 3 | 0.906716 | 0.517075 | 2.171259 | 9.249501 | 16.077464 |
| 4 | 2.813201 | 1.608974 | 1.827349 | 7.380719 | 11.498883 |
| ... | ... | ... | ... | ... | ... |
| 9211 | 4.055669 | 3.663706 | 1.711899 | 6.189053 | 10.337699 |
| 9212 | 2.986643 | 0.230117 | 1.837787 | 8.302855 | 11.645122 |
| 9213 | 5.124904 | 3.761257 | 1.732857 | 7.683230 | 12.600224 |
| 9214 | 3.533581 | 3.544352 | 1.821830 | 7.305826 | 9.119884 |
| 9215 | 3.413049 | 1.736568 | 1.751540 | 6.413657 | 11.491378 |
9216 rows × 5 columns